Protein structures
Krzysztof Brzozowski
University of Gdansk, Faculty of Chemistry
Proteins is a group of biopolymers which has attracted a lot of attention for quite a long time. They are often modified by phosphorylation, glycosylation or acylation what influences the polymers’ regulatory, metabolic or physiological activity. That is why understanding of Structure-Activity Relationship (SAR) is one of the tasks of structural biochemistry. Knowledge of the structural details allows to understand the molecular mechanism of action, and dynamic behaviour of the molecule under different conditions.
One of the classical examples of choosing separation parameters is pH optimization. This value is connected with isoelectric point (pI) what is related with the form the molecule exists (cationic, anionic or zwitterion). Figure 1 shows the relationship between pI and pH.
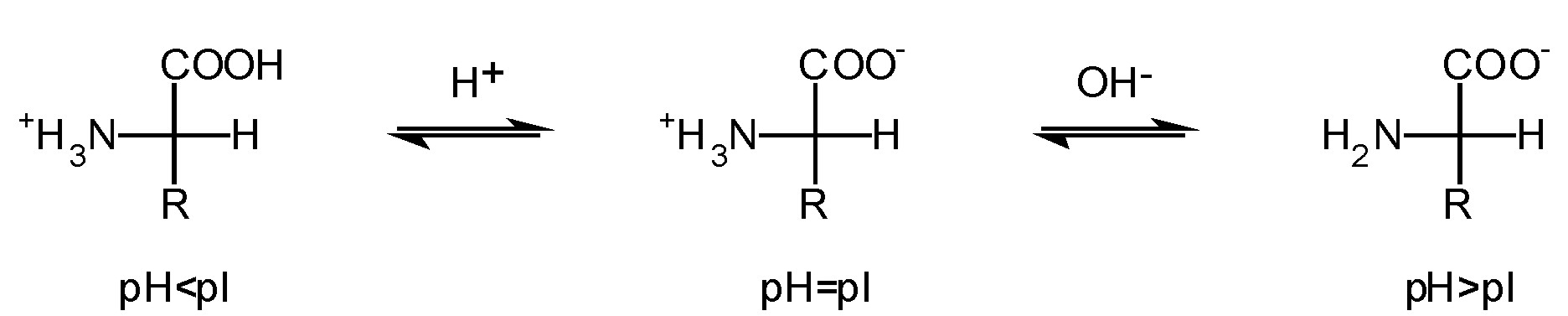
Figure 1. Dependency between pH and pI

Figure 1. Dependency between pH and pI
Proteins consist mainly of 20 common amino acids. Non-natural amino acids which may be present in the sequence are introduced during post-translational processing. Their side chains have different shapes, electrostatic charge and chemical activity. Moreover they display different tendency towards hydrogen bonds formation.
Taking the chemical character of amino acids into account, they can be divided into the following categories:
- acidic: Asp, Asn, Glu, Gln
- basic: Lys, Arg, His
- aromatic: Phe, Trp, Tyr
- containing –OH groups or S atoms: Ser, Thr, Cys, Met
- aliphatic: Gly, Ala, Val, Leu, Ile, Pro
In tables 1 and 2 common and non-natural amino aid residues are shown respectively.
Table 1. Common amino acid residues

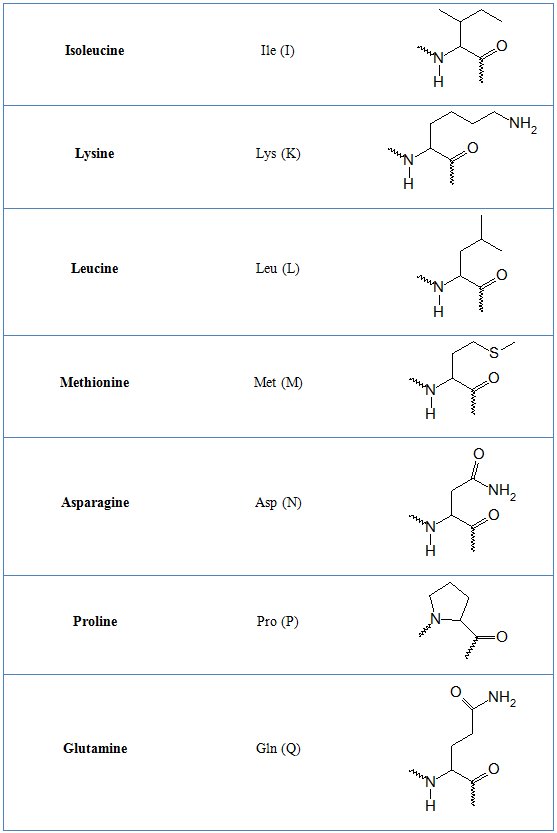
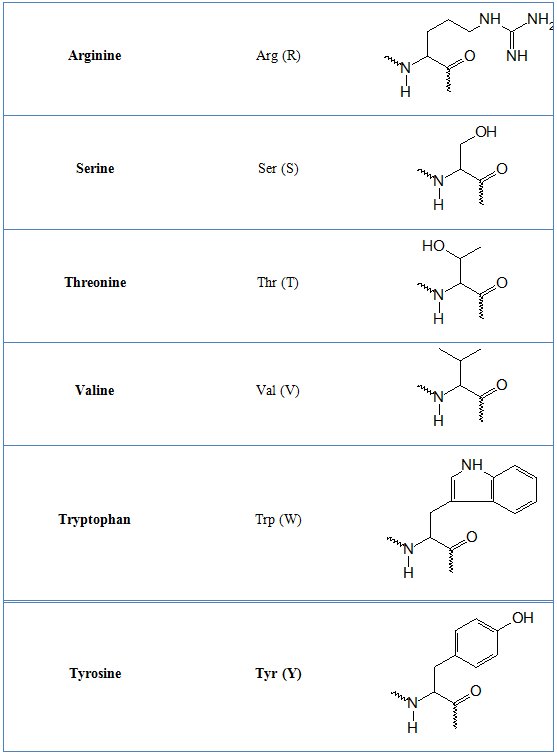
Table 2. Non-standard amino acid residues
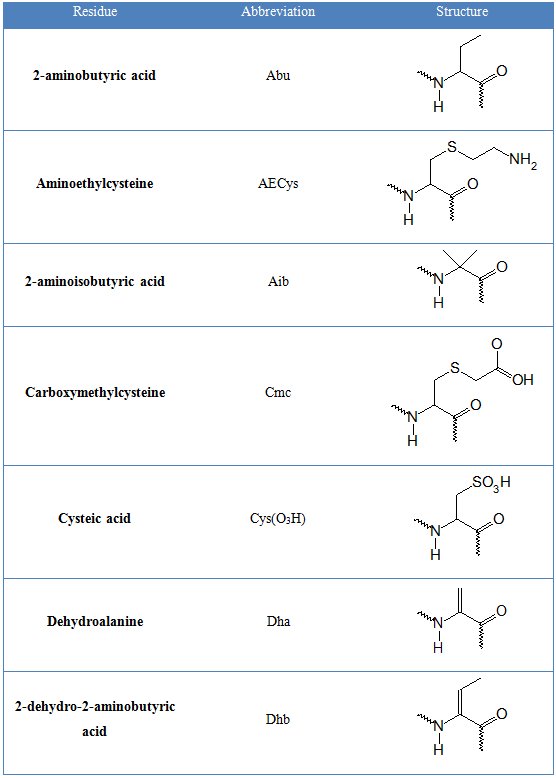
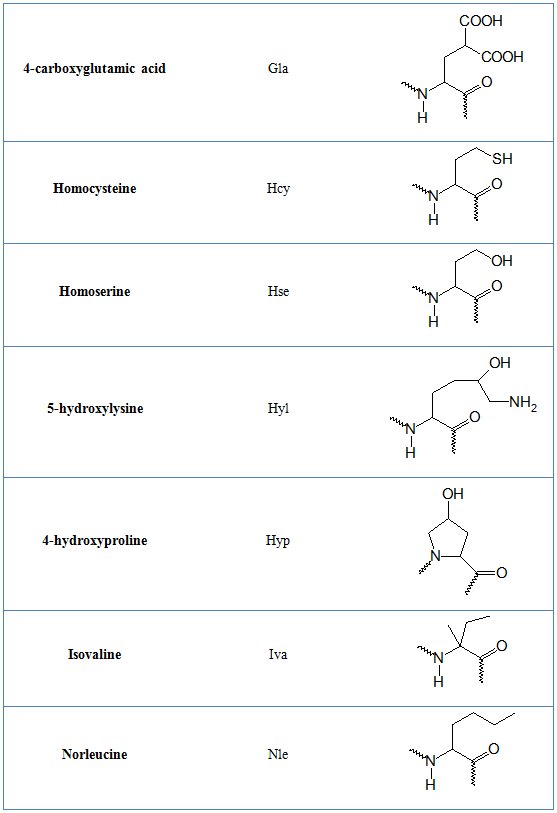

Table 1. Common amino acid residues



Table 2. Non-standard amino acid residues



General structural information
Amino acid sequence and placement of disulphide bridges in polypeptide chain is called primary structure. Such chains can form more complex, three dimensional secondary structure elements, namely helices or β-sheets. Their mutual arrangement is tertiary structure and quaternary structure is interaction of single proteins leading to formation of dimers, trimers, etc. According to Chou and Fasman, there are α-helix and β-sheet promoters, neutral and structure breaker amino acids – table 3 [1].
Table 3. Tendency of amino acid residues for promoting and breaking secondary structures
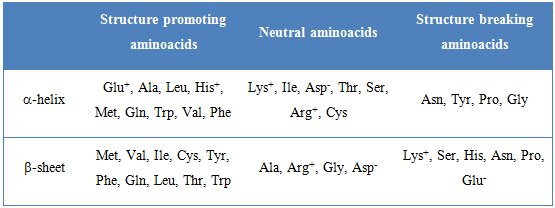
Table 3. Tendency of amino acid residues for promoting and breaking secondary structures

The three dimensional structure of folded protein is stabilized by factors displayed in figure 2.

Figure 2. Interactions stabilizing structures of proteins. Based on [2]

Figure 2. Interactions stabilizing structures of proteins. Based on [2]
Secondary structure
Helix
The helix is very similar to left or right handed screw. There are several types of this structural element and regardless of the type it is stabilized by the hydrogen bond. Helices are defined by two main parameters – Nn, where N is the number of amino acid residues per turn and n is the number of backbone atoms closed by hydrogen bond. Giving whole characteristics some additional information such as the helix pitch (repeat distance – 0.54 nm for α-helix) and the distance between neighbouring amino acid residues (0.15 nm for α-helix) can be provided [2]. Figure 3 shows the representation of three types of helices and table 4 presents their structural characteristics.
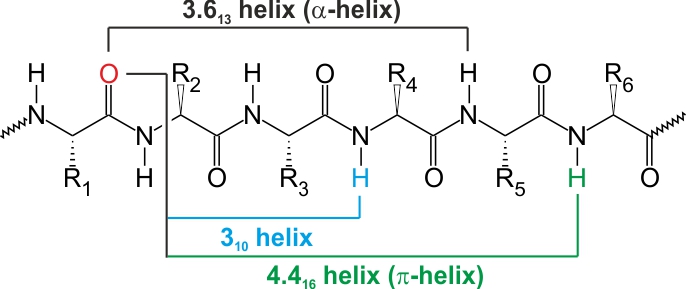
Figure 3. The schematic of different types of helices. Based on [2]
Table 4. The features of different types of helices


Figure 3. The schematic of different types of helices. Based on [2]
Table 4. The features of different types of helices

β-sheet
Contrary to previously described structural element some amino acids do not have any ability to form intra-chain hydrogen bonds. Instead the three dimensional structure is stabilized by inter-chain interactions. It results with extended structure called β-pleated sheet or β-sheet. Comparing to helix the distance between neghbouring amino acid residues is bigger and equal to 0.72 nm. The values of ϕ and ψ are -139° and 135° for antiparallel and -119° and 113° for parallel β-sheet respectively. Examples of both structures are shown in figure 4 [3].
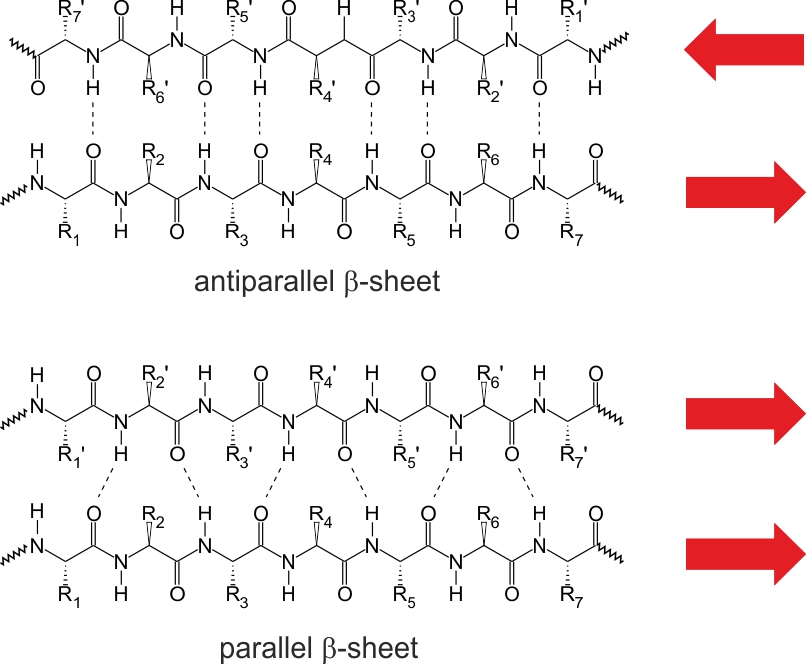
Figure 4. Two types of β-sheet structure. Based on [3]

Figure 4. Two types of β-sheet structure. Based on [3]
Turns
β and γ are the most common turns present in protein structures – figure 5 and table 5 [2]. They involve four and three amino acid residues respectively. There are also α and π turns formed by five and six amino acid residues respectively. The result of turns presence is tight folding and globular shape of the molecule.
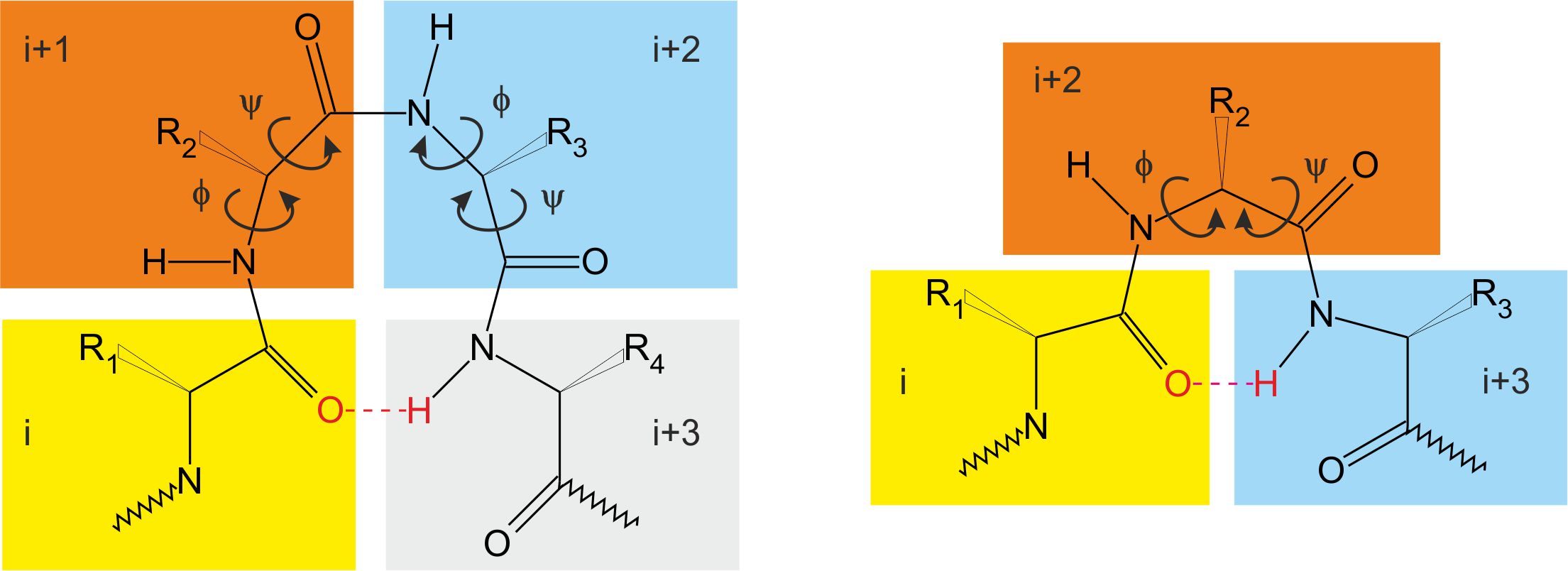
Figure 5. β (left) and γ (right) turns. Based on [2]
Table 5. The geometry of β and γ turns [2]
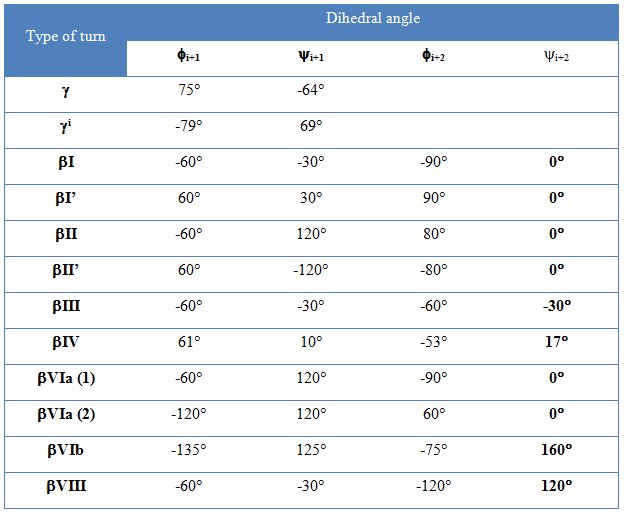

Figure 5. β (left) and γ (right) turns. Based on [2]
Table 5. The geometry of β and γ turns [2]

Structural domains in proteins
Analysis of many protein structures revealed the existence of some common motifs called structural domains. They are formed by certain location of secondary structure elements. The domains are also responsible for biological properties of these biopolymers. Below there is a short description of common domains located in protein molecules.
α-helix
It is particularly the most common element. Helices containing proteins can be either globular – figure 6 [4] or fibrillar – figure 7 [5] in shape. The key structural domain element of membrane receptors is leucine zipper shown in figure 8 [6].
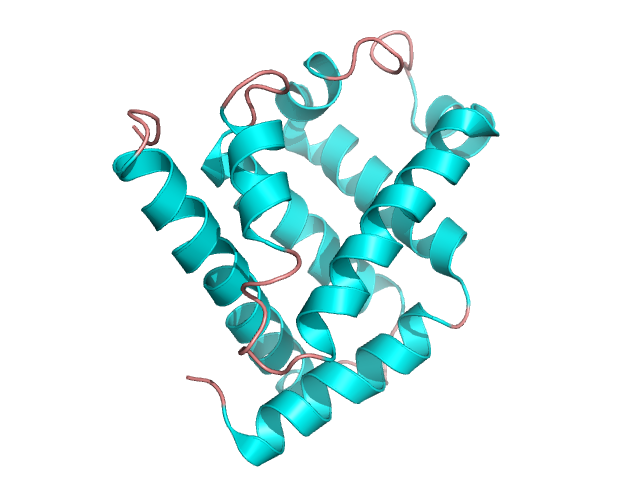
Figure 6. The structure of myoglobin. Based on pdb [1mbn] [4]
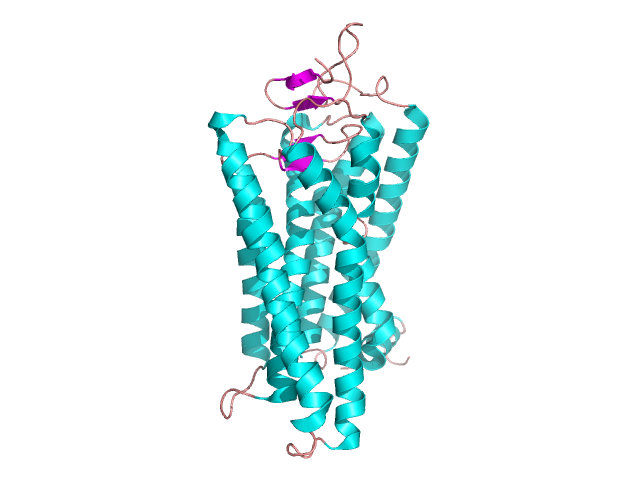
Figure 7. The structure of bovine rhodopsin. Based on pdb [1gzm] [5]
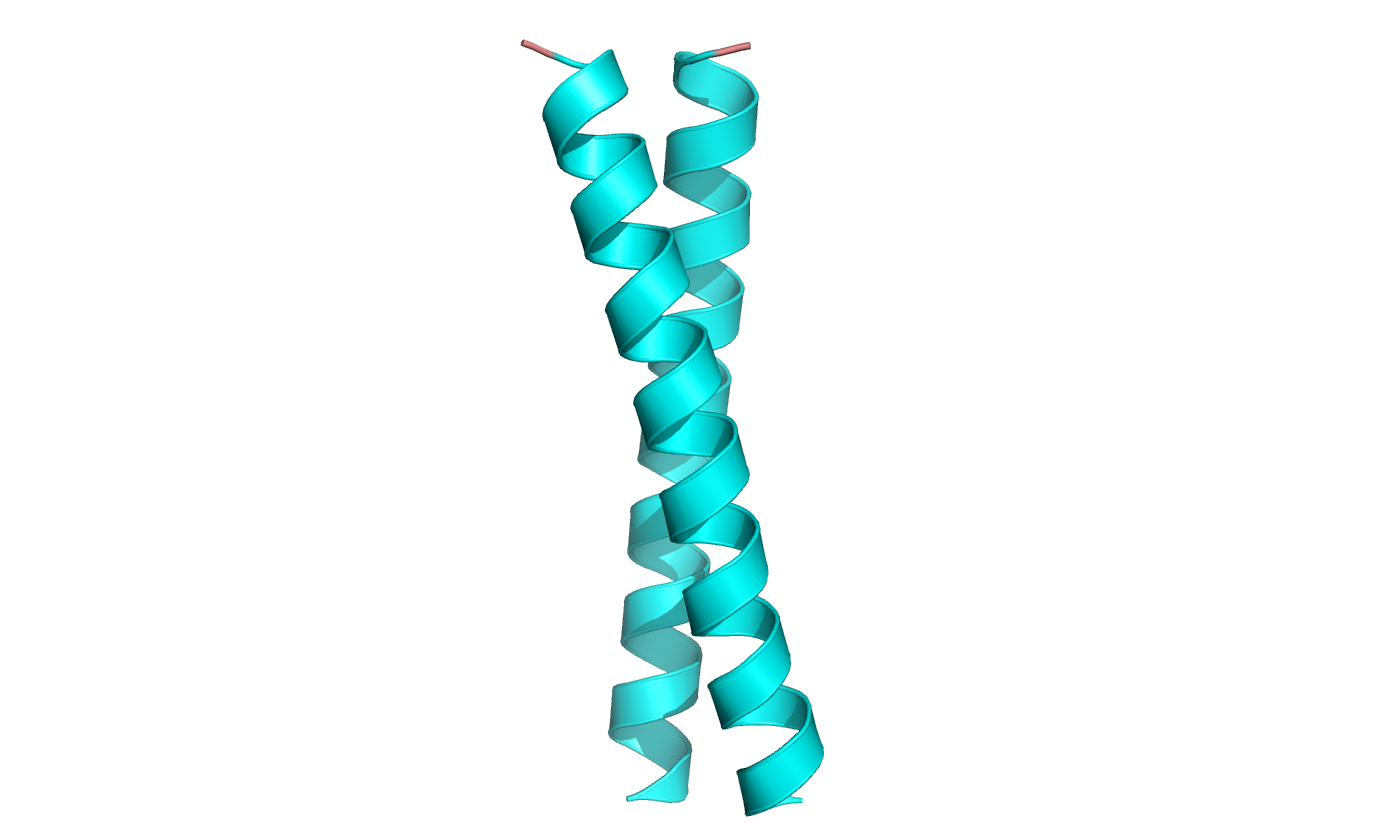
Figure 8. The leucine zipper – an element of transmembrane receptors. Based on pdb [1zij] [6]

Figure 6. The structure of myoglobin. Based on pdb [1mbn] [4]

Figure 7. The structure of bovine rhodopsin. Based on pdb [1gzm] [5]

Figure 8. The leucine zipper – an element of transmembrane receptors. Based on pdb [1zij] [6]
Among all examples of helices, collagen is a unique one. This protein consists of three 1000 amino acid residues long polypeptide chains – figure 9 [7]. Their amino acid sequence involves a repeating unit of –Gly-Pro-Hyp-. All strands are stabilized by inter-chain hydrogen bonds and pyrrolidic rings of Pro and Hyp do not cause any steric hindrance.
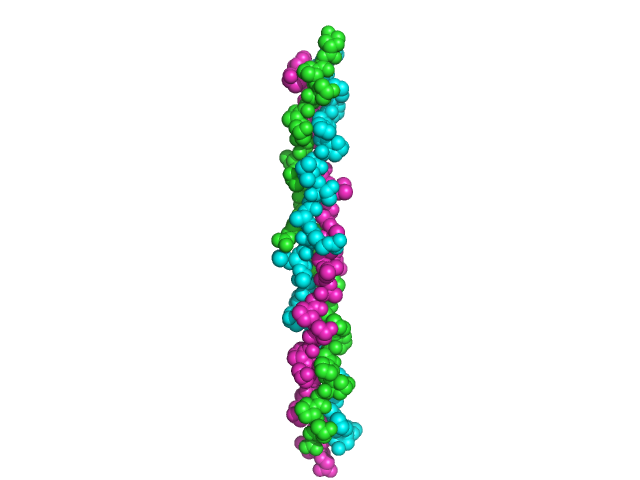
Figure 9. Collagen structure. Based on pdb [1bkv] [7]

Figure 9. Collagen structure. Based on pdb [1bkv] [7]
βαβ
This domain is a combination of two β-sheets and one α-helix – figure 10. Such a structure is present in one of dehydrogenases forming TIM barrel – figure 11 [8].
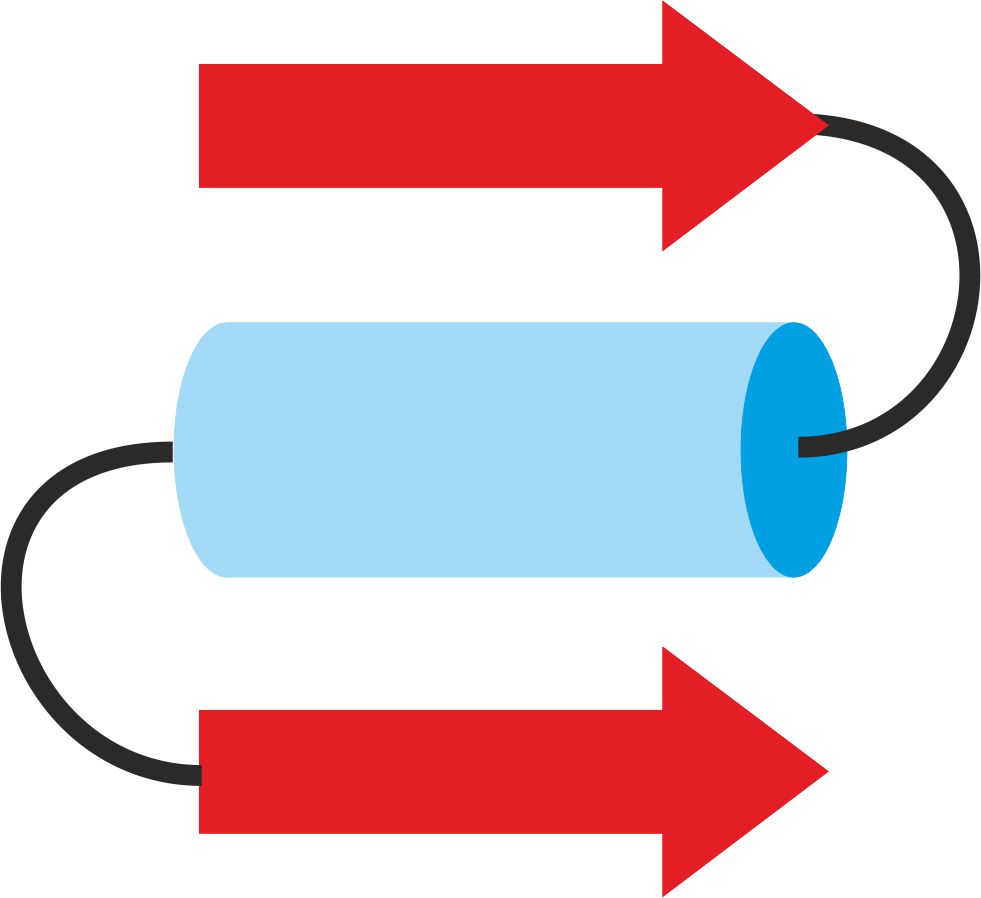
Figure 10. βαβ domain
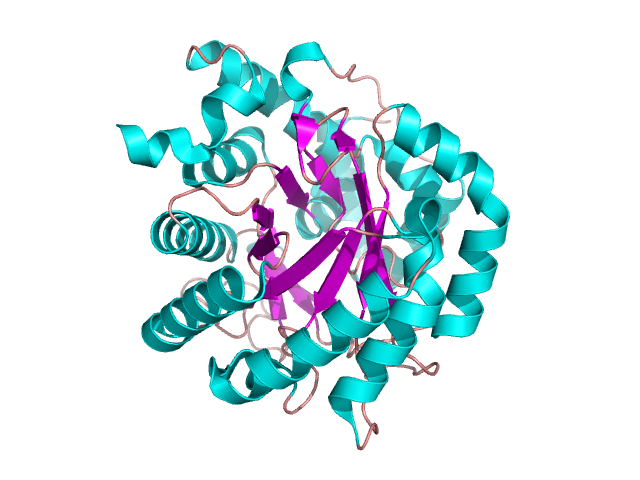
Figure 11. The structure of dihydroorotate dehydrogenase. Based on pdb [2wv8] [8]

Figure 10. βαβ domain

Figure 11. The structure of dihydroorotate dehydrogenase. Based on pdb [2wv8] [8]
β-sheets domains
A lot of domain structures are formed by different arrangement of β-pleated sheets – figure 12. The particular examples of proteins containing them are given in the following pictures: the Greek key – figure 13 [9], β-sandwich –figure 14 [10], β-barrel – figure 15 [11], β-propeller – figure 16 [12] and β-saddle – figure 17 [13]. The first domain in stabilized by a connection between two edge sheets in four strand unit. β-sandwich is a multi-level structure where upper strands are connected with lower ones. Other mentioned structures are typical antiparallel β-sheets with different arrangements depending on biological properties.
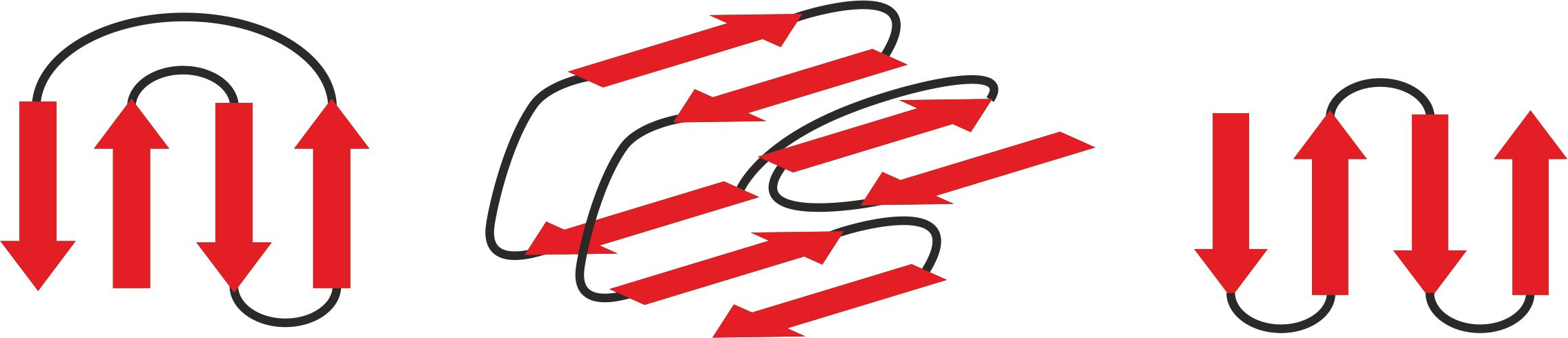
Figure 12. The backbones of three β-sheet domains: the Greek key (left), β-sandwich (middle), antiparallel β-sheet (right)
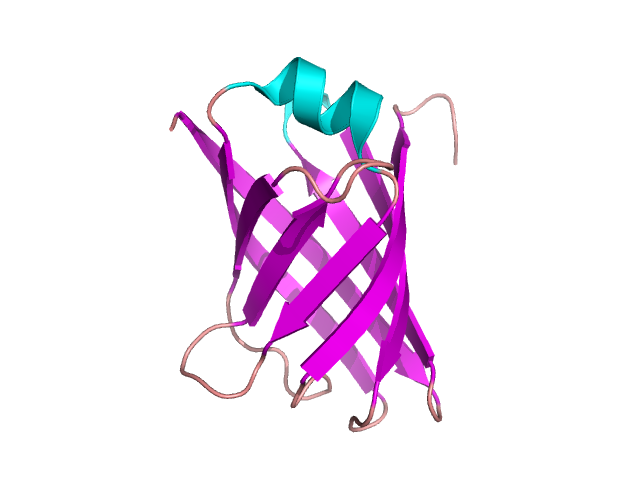
Figure 13. The structure of VAPB protein containing the Greek key motif. Based on pdb [4cv7] [9]
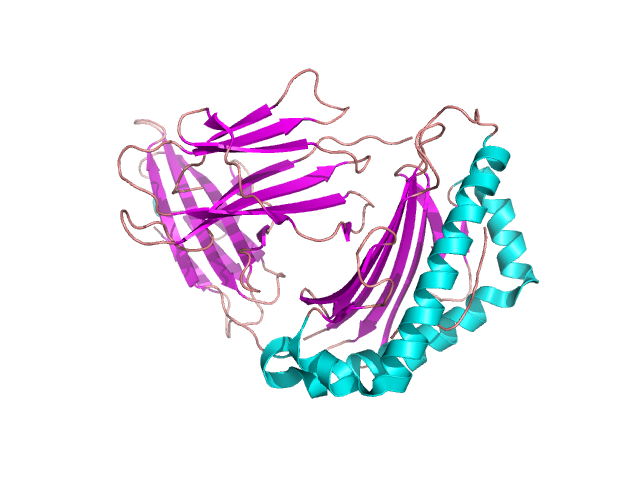
Figure 14. β-sandwich structure in β-2-globulin. Based pdb [3bgm] [10]
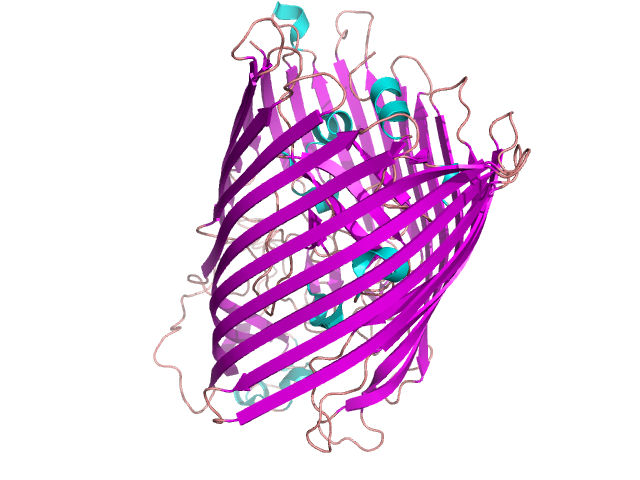
Figure 15. FHUA protein containing β-barrel domain. Based on [1fi1] [11]
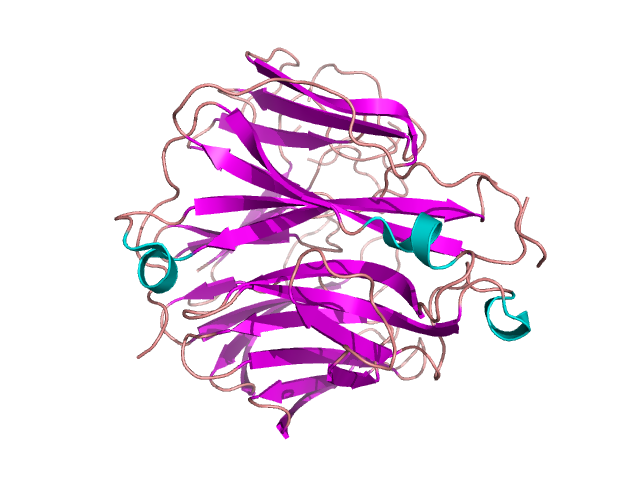
Figure 16. β-propeller in oseltamivir-resistant influenza virus neuromidase mutant. Based on [3c10] [12]
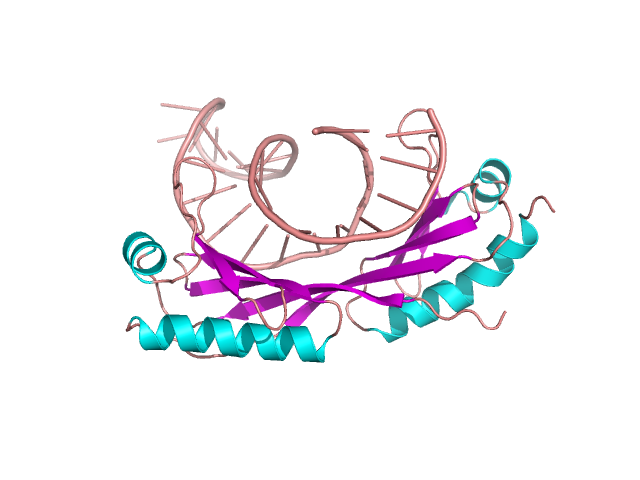
Figure 17. DNA and TATA protein complex – example of β-saddle domain. Based on [1ytb] [13]

Figure 12. The backbones of three β-sheet domains: the Greek key (left), β-sandwich (middle), antiparallel β-sheet (right)

Figure 13. The structure of VAPB protein containing the Greek key motif. Based on pdb [4cv7] [9]

Figure 14. β-sandwich structure in β-2-globulin. Based pdb [3bgm] [10]

Figure 15. FHUA protein containing β-barrel domain. Based on [1fi1] [11]

Figure 16. β-propeller in oseltamivir-resistant influenza virus neuromidase mutant. Based on [3c10] [12]

Figure 17. DNA and TATA protein complex – example of β-saddle domain. Based on [1ytb] [13]
Zinc finger
This structural element is present in DNA binding proteins. It consists of 30 amino acids forming two antiparallel β-sheet strands and one α-helix. The presence of Zn2+ is crucial for stabilization of the domain. It is chelated by two His and two Cys residues – figure 18 [14].
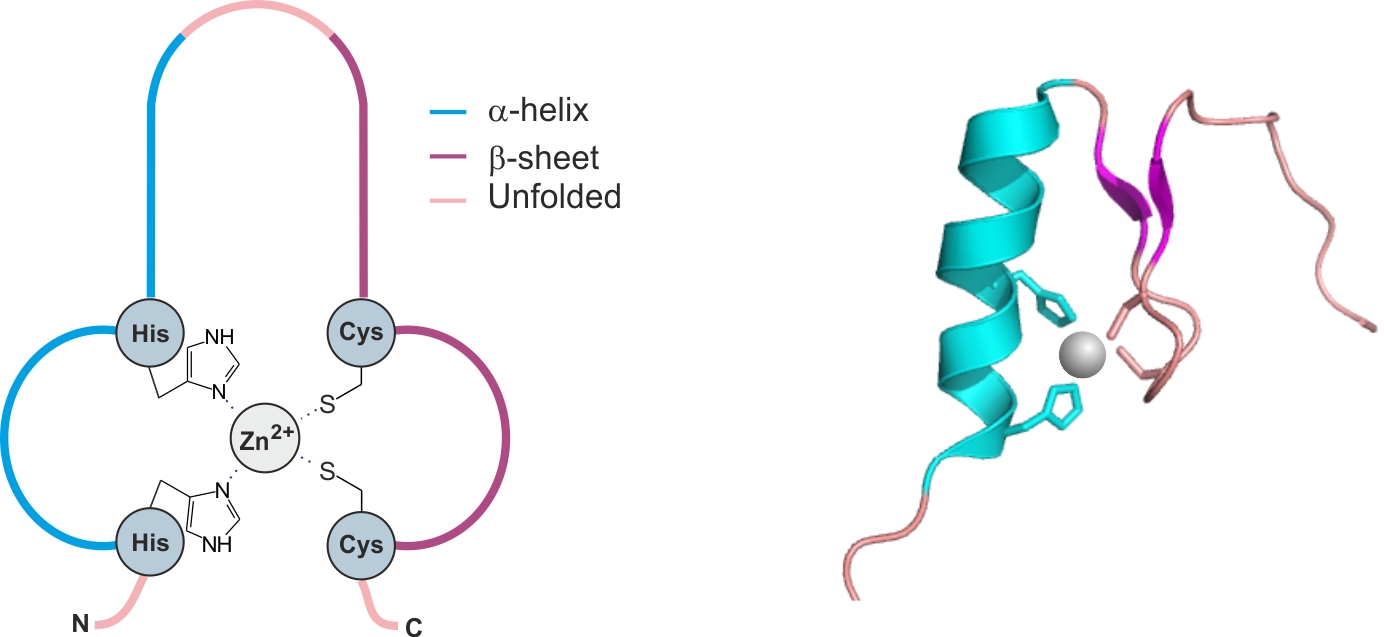
Figure 18. The zinc finger domain. Based on [1x5w] [14]

Figure 18. The zinc finger domain. Based on [1x5w] [14]
The conformational complexity of proteins is not a closed matter. Searching the database one can find countless amount of structures containing elements described above. The bright side of such diversity is a guarantee of work for structural chemists for a long time.
References
[1] G.C. Barret, D.T. Elmore. Amino Acids and Peptides, Cambridge University Press, UK (2004).
[2] N. Sewald, H.D. Jakubke. Peptides: Chemistry and Biology, Wiley-VCH Verlag GmbH (2002).
[3] J.M. Berg, J.L. Tymoczko, L. Stryer. Biochemistry, Fifth Edition, W.H. Freeman, New York (2002).
[4] H.C. Watson, The Stereochemistry of the Protein Myoglobin, Prog. Stereochem., 4, 299 (1969).
[5] J. Li, P. Edwards, M. Burghammer, C. Villa, G.F.X. Schertler, Structure of Bovine Rhodopsin in a Trigonal Crystal Form, J. Mol. Biol., 343, 1409 (2004).
[6] E.K. O’Shea, J.D. Klemm, P.S.Kim, T. Alber, X-Ray Structure of the GCN4 Leucine Zipper, a Two Stranded, Parallel Coiled Coil, Science, 254, 539 (1991).
[7] R.Z. Kramer, J. Bella, P. Mayville, B. Brodsky, H.M. Berman, Sequence Depended Conformational Variations of Collagen Triple-Helical Structure, Nat. Struct. Biol., 6, 454 (1999).
[8] I. Fritzson, B. Svenson, S.Al-Karadaghi, B. Walse, U. Wellmar, U.J. Nillsson, D. Da GracaThrige, S. Johnson, Inhibition of Human Dhoby by 4-Hydroxycoumarins, Fenamic 2 Acids, and N-(Alkylcarbonyl)anthranilic Acid Identified by Structure-Guided Fragment Selection, Chemmedchem, 5, 608 (2010).
[9] C. Geerds, J. Wohlmann, A. Haas, H.H. Niemann, Structure of Rhodococcus Equi Virulence-Associated Protein B (VAPB) Reveals an Eight-Stranded Antiparallel [Beta]-Barrel Consisting of Two Greek-Key Motifs, Acta Crystallogr., Sect. F, 70, 866 (2014).
[10] F. Mohammed, M. Cobbold, A.L. Zarling, M. Salim, G.A. BarrettiWilt, J. Shabanowitz, D.F. Hunt, V.H. Engelhard, B.E. Willcox, Phosphorylation-Dependent Interaction Between Antigenic Peptides and MHC Class I: A Molecular Basis for the Presentation of Transformed Self, Nat. Immunol., 9, 1236 (2008).
[11] A.D. Ferguson, J. Kodding, G. Walker, C.Bos, J.W. Coulton, K. Diederichs, V. Braun, W. Welte, Active Transport of an Antibiotic Rifsmycin Derivative by the Outer-Membrane Protein FHUA, Structure, 9, 707 (2001).
[12] P.J. Collins, L.F. Haire, Y.P. Lin, J. Liu, R.J. Russell, P.A. Walker, J.J. Skehel, S.R., Martin, A.J. Hay, S.J. Gamblin, Crystal Structures of Oseltamivir-Resistant Influenza Virus Neuramidase Mutants, Nature, 453, 1258 (2008).
[13] Y. Kim, J.H. Geiger, S. Hahn, P.B. Sigler, Crystal Strycture of a Yeat TBP/TATA-Box Complex, Nature, 512, (1993).
[14] M. Yoneyama, S. Koshiba, N. Tochio, M. Inoue, T. Kigawa, S. Yokoyama, Solution Structure of the C2H2Type Zinc-Binding Domain of Human Zinc Finger Protein 64, Isoforms 1 and 2 w druku.
References
[1] G.C. Barret, D.T. Elmore. Amino Acids and Peptides, Cambridge University Press, UK (2004).
[2] N. Sewald, H.D. Jakubke. Peptides: Chemistry and Biology, Wiley-VCH Verlag GmbH (2002).
[3] J.M. Berg, J.L. Tymoczko, L. Stryer. Biochemistry, Fifth Edition, W.H. Freeman, New York (2002).
[4] H.C. Watson, The Stereochemistry of the Protein Myoglobin, Prog. Stereochem., 4, 299 (1969).
[5] J. Li, P. Edwards, M. Burghammer, C. Villa, G.F.X. Schertler, Structure of Bovine Rhodopsin in a Trigonal Crystal Form, J. Mol. Biol., 343, 1409 (2004).
[6] E.K. O’Shea, J.D. Klemm, P.S.Kim, T. Alber, X-Ray Structure of the GCN4 Leucine Zipper, a Two Stranded, Parallel Coiled Coil, Science, 254, 539 (1991).
[7] R.Z. Kramer, J. Bella, P. Mayville, B. Brodsky, H.M. Berman, Sequence Depended Conformational Variations of Collagen Triple-Helical Structure, Nat. Struct. Biol., 6, 454 (1999).
[8] I. Fritzson, B. Svenson, S.Al-Karadaghi, B. Walse, U. Wellmar, U.J. Nillsson, D. Da GracaThrige, S. Johnson, Inhibition of Human Dhoby by 4-Hydroxycoumarins, Fenamic 2 Acids, and N-(Alkylcarbonyl)anthranilic Acid Identified by Structure-Guided Fragment Selection, Chemmedchem, 5, 608 (2010).
[9] C. Geerds, J. Wohlmann, A. Haas, H.H. Niemann, Structure of Rhodococcus Equi Virulence-Associated Protein B (VAPB) Reveals an Eight-Stranded Antiparallel [Beta]-Barrel Consisting of Two Greek-Key Motifs, Acta Crystallogr., Sect. F, 70, 866 (2014).
[10] F. Mohammed, M. Cobbold, A.L. Zarling, M. Salim, G.A. BarrettiWilt, J. Shabanowitz, D.F. Hunt, V.H. Engelhard, B.E. Willcox, Phosphorylation-Dependent Interaction Between Antigenic Peptides and MHC Class I: A Molecular Basis for the Presentation of Transformed Self, Nat. Immunol., 9, 1236 (2008).
[11] A.D. Ferguson, J. Kodding, G. Walker, C.Bos, J.W. Coulton, K. Diederichs, V. Braun, W. Welte, Active Transport of an Antibiotic Rifsmycin Derivative by the Outer-Membrane Protein FHUA, Structure, 9, 707 (2001).
[12] P.J. Collins, L.F. Haire, Y.P. Lin, J. Liu, R.J. Russell, P.A. Walker, J.J. Skehel, S.R., Martin, A.J. Hay, S.J. Gamblin, Crystal Structures of Oseltamivir-Resistant Influenza Virus Neuramidase Mutants, Nature, 453, 1258 (2008).
[13] Y. Kim, J.H. Geiger, S. Hahn, P.B. Sigler, Crystal Strycture of a Yeat TBP/TATA-Box Complex, Nature, 512, (1993).
[14] M. Yoneyama, S. Koshiba, N. Tochio, M. Inoue, T. Kigawa, S. Yokoyama, Solution Structure of the C2H2Type Zinc-Binding Domain of Human Zinc Finger Protein 64, Isoforms 1 and 2 w druku.


